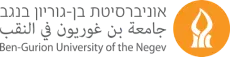Research
Researcher: Dr. Assaf Zaritsky
(computational cell dynamics, data science in cell imaging)
Student: Katya Smoliansky
Using Microscopy and Generative Networks to Infer Novel Molecular Interactions PI: Assaf Zaritsky (computational cell dynamics, data science in cell imaging) Department of Software and Information Systems Engineering, Ben-Gurion University of the Negev, Beer Sheva 84105, Israel.
Email: assafzar@gmail.com. Phone: 972-50-753-8585.
Lab website
Cells are the fundamental unit of structure and function of all organisms. Proteins are the molecular machines that define the cell architecture, organization, and function. Microscopy is the only technology that allows us to see live cell behavior and to correlate cell function to protein quantity and location. Recent studies have demonstrated that label-free (without fluorescent stains for specific molecules) images contain information on the molecular organization within the cell by using machine learning-based generative approaches.
The potential generation of such “virtual integrated cells” may overcome inherent limitations in optics and molecular biology that limits the number of proteins that can be concurrently images in a live cell. We propose to make the next big step by using matched fluorescent and label free images to predict (asymmetric) protein-protein interactions that will be used to identify novel molecular pathways that can be then verified experimentally. Interactions among different proteins is poorly characterized due to the difficulty in live imaging multiple different proteins in a cell.
We propose here a proof-of-concept of a methodology that will enable predictive modeling of cell states and behavior, based on virtual predicted molecular organization and interactions from label free images. Such system-level understanding of molecular interactions will be a huge advance towards the “holy grail” of cell biology - an understanding of the cell as an integrated complex system.
Students taking part in this project: Katya Smoliansky, M.Sc. student, Department of Computer Science. Gil Baron, M.Sc. student, Department of Software and Information Systems Engineering.  Summary
Summary
Researchers: Dr. Tirza Routtenberg (School of Electrical and Computer Engineering), Dr. Jonathan D. Rosenblatt (Department of Industrial Engineering and Management)
Student: Neta Zimerman
Estimation of the direction of arrival (DOA) of a seismic signal is required for accurate localization of events such as earthquakes and human-made explosions. While seismic noise has very low amplitude and cannot be felt by humans, it has a significant influence o the accuracy of DOA. Currently, most seismic DOA estimation algorithms are based on the assumption that the additive seismic noise is uncorrelated between sensors. However, in our preliminary results we show by analyzing real-data sets that seismic sensors exhibit noise correlation, and thus, the uncorrelated assumption results in poor performance in terms of localization of seismic events. In order to compensate for this correlated noise we develop new tools and algorithms for DOA and array design that are based o robust statistics, array signal processing, and optimization theory, and account for different noise types.
Publication: N. Zimerman, J. D. Rosenblatt, and T. Routtenberg, "Colored Noise in DOA Estimation from Seismic Data: an Empirical Study," Submitted to Asilomar Conference on Signals, Systems, and Computers 2020.
Researchers: Prof. Aryeh Kontorovich, Computer Science -- statistical learning, big data and Dr. Rami Puzis, Software and Information Systems Engineering -- network analysis, cyber security
Students: דורון כהן, טטיאנה פרנקלך
Smartphones are an essential part of our everyday life, providing a means of communication, information, self-care services, entertainment, and more. Yet, Android app markets are filled with numerous malicious applications that threaten mobile users' security and privacy. Due to the ever-increasing number of both malicious and benign applications, there is a need for scalable malware detectors that can efficiently address big data challenges. Motivated by large-scale recommender systems, we propose a static application analysis method that differentiates malicious and benign applications based on an app similarity graph (ASG) and classification based on nearest neighbors. In order to address the big-data challenges, we perform sample compression where a small number of significant anchor APKs are selected in order to represent the complete dataset.
We demonstrate our method on the Drebin benchmark in both balanced and unbalanced settings and on a dataset of approximately 190k applications provided by VirusTotal, achieving an accuracy of 0.975 in balanced settings.
Researchers: Dr. Eran Treister, Computational Mathematics. Department of Computer Sciences, Faculty of Natural Sciences and Dr. Tal Shay, Computational Biology and Immunoinformatics Department of Life Sciences, Faculty of Natural Sciences.
Student: סופי פלדמן
Researcher: Prof. Daniel Berend – Applied Probability, Combi- natorial Optimization.
SAT is one of the most celebrated problems in Computer Science. It is of both theoretical and practical importance.
MAX-SAT is a combinatorial optimization version of SAT.
The proposers have worked for several years on finding good heuristics for MAX-SAT, that may have analogues for other combinatorial optimization problems. In this proposal, they suggest ways for employing ML techniques to these problems.
Researcher: Prof. Lior Rokach
Researchers: Prof. Mark Last: Natural Language Processing, Machine Learning; Dr. Armin Shmilovici: Data Mining, Expert Systems, Stochastic Modeling;
video clips taken from 199 movies, covers the entire affect range. The clips range from calm conversation to intense action sequences. For discrete emotion categories Cowen and Keltner’s (2017) consists of 2,185 emotion-labeled clips dataset, while Chu, (2017) consist of 1,000 video clips.
Various audio-visual features are used for predicting the emotions induced by video-clips. These include low-level statistical features such as audio and color histograms and energy statistics (Mo et al. ,2018), high level features such as audio-visual words (Irie et al., 2010), non-linear transformations of the audio and video data such as the Hilbert–Huang Transform (Mo et al., 2018), and output of a pre-trained deep neural network such as the C3D encoding (Carreira, 2018). However, to the best of our knowledge, no one has evaluated the contribution of textual video descriptions to the emotion prediction task.
Research Methodology: Initially, we will establish a baseline using multiple regression schemes of low-level audio-visual statistical features, and determine the most relevant features. Than we will construct emotion identification systems based on high-level features such as automatically generated textual descriptions of video-clips (Miech et al, 2019). We will train the low-level regressors as well as high-level feature regressors on various datasets of annotated video clips (such as LIRIS-ACCEDE) and will evaluate them on datasets of full videos such as the 51 movies of Vicol et al. (2018). The primary metric for evaluation of emotion identification methods is Concordance Correlation Coefficient (CCC) between the annotated ground truth and the predicted ratings on the test set. This metric accounts for both the similarity of pattern between the two-time series as well as the bias difference between them.
Specific Research Goals: We plan to develop machine-learning methods to identify the main emotion-invoking elements of a story in a movie and their expected emotional effect (such as fear or tenderness) on a typical human viewer.
Researchers: Dr. Arnon Sturm and Dr. Roni Stern, Department of Software and Information Systems Engineering
Software developers nowadays handle tasks that involve multiple programming languages, databases, and frameworks. To utilize these diverse set of development tools, developers often search open information sources such as Stackoverflow and GitHub. Yet, searching these sources require significant efforts, since existing search tools and engines for these information sources support limited types of searches, mainly based on keywords matching. In this research, we address this challenge by developing a search framework that maps the existing software knowledge into a predefined schema that have a simple semantic and enables powerful sematic search capabilities. The impact of such an intelligent and comprehensive search framework is expected to be transformative, significantly simplifying and shortening the searching time and making it more effective, thereby lowering software development cost and time and improving its quality.
Researcher: Prof. Bracha Shapira
Multimodal machine learning deals with building models that can process information from multiple modalities. A modality refers to a way of doing or experiencing something. Examples of modalities processed in machine learning models include: video, audio, textual data, and spatial data. Multimodal machine learning is gaining research interest due to its relevance to many fields where in most cases, the research focuses on unstructured data such as video, audio, and text.
The main objective of the proposed research is to develop novel methods for multimodal learning utilizing representations created from different data types (modalities). To pursue this objective, we present a series of relevant modeling tasks.
We intend to define each of the tasks formally, design and implement scalable models and algorithmic solutions based on machine learning and similar methods, and finally, evaluate the models using standard methods.
Drug research and development as well as guaranteeing drug safety are complex and expensive tasks, which usually require experiments involving humans. On the other hand, several drug-related data sources are available in various modalities. One example is DrugBank which contains different types of structured data: interaction graph, tabular data, molecular structures and more. For these two reasons, we intend to use drug knowledge to asses the models proposed in this research. We present a series of drug-related tasks which will allow us to evaluate the models proposed during this research and to discover and contribute new drug knowledge.
The results from the preliminary research are encouraging. Several experiments were performed: during the first experiment series we predicted drug-interaction using a novel method for link prediction in graphs. Additionally, we showed that known drug-interactions are the preferred data source for predicting new drug-interactions compared to nineteen data sources. The second experiment series dealt with creating embeddings using medical ontology. The results showed that word embeddings created over medical ontology can improve named entity recognition performance. The last experiment series dealt with classifying drugs that are safe for usage during pregnancy. The results demonstrate that by analyzing pharmacologist's and doctor's answers to queries from the general public, drug safety during pregnancy can be classified with an area under the receiver operating characteristics curve score of 98\%. These results encourage us to continue the research by predicting additional drug-related information and expanding it to new modeling tasks. These preliminary results lead to a publication in PLOS One.
Researcher: Dr. Achiya Elyasaf, Collaborator: Mr. Amit Livne
user context to provide personalized services. The contextual information can be driven from sensors in order to improve the accuracy of the recommendations. Yet, generating accurate recommendations is not enough to constitute a useful system from the users' perspective, since certain contextual information may cause different issues, such as draining the user's battery, privacy issues, and more. Additionally, adding high-dimensional contextual information may increase both the dimensionality and sparsity of the model.
Previous studies suggest reducing the amount of contextual information by selecting the most suitable contextual information using domain knowledge. While in most studies the set of contexts is both small enough to handle and sufficient to prevent sparsity, such context sets do not necessarily represent an optimal set of features for the recommendation process. Another solution is compressing it into a denser latent space, thus disrupting the ability to explain the recommendation item to the user, and damaging users' trust.
The main goal of this proposal is to develop new methodologies for creating CARS models, that will allow for controlling user aspects, such as privacy and battery consumption. Specifically, we propose to develop a feature-selection algorithm, based on evolutionary algorithms (EA), for explicitly incorporating contextual information within CARS (see Figure 1). In a preliminary experiment with a small CARS dataset derived from mobile phones, EA produced different subsets of contextual features that demonstrated similar high-accuracy values and interoperability of the provided recommendations. Thus, we were able to manually examine the effect of the selected features on both accuracy and user aspects (i.e., privacy and battery optimization).
We believe that our approach will contribute to the emerging field of explainable dynamic CARS, and yield a novel type of explanations. Moreover, this approach can be later extended to general recommender systems and contribute to the larger community.
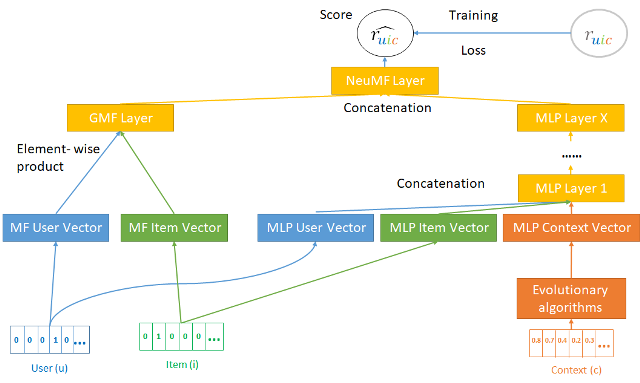
Figure 1. The proposed framework. An evolutionary algorithm evolves subsets of the contextual information (features), that are explicitly incorporated within a context-aware recommender system.
Researchers: Dr. Yaniv Zigel, Biomedical Engineering, Prof. Haim Permuter, Electrical Engineering. Collaborator: Prof. Nimrod Maimon, MD, heads the Medicine B and Corona B departments, Soroka Medical Center.
In the current COVID-19 disease pandemic, pneumonia appears to be the most frequent serious manifestation of infection. It is characterized primarily by fever, cough, dyspnea, and an excess of respiratory airway secretions, which may be recognized automatically using audio analysis techniques. Early diagnosis and treatment may curb disease spread and ameliorate its management, for instance by conducting remote screening of asymptomatic or symptomatic carriers, isolated communities or hospitalized patients to achieve early detection and to perform risk stratification for predicted clinical deterioration.
In this research, we introduce a novel and original technological concept to diagnose COVID-19 from audio signals, including: 1) speech, 2) breathing, and 3) coughing using a simple audio recorder (e.g. smartphone). We aim to explore and develop signal processing and machine learning algorithms for the detection of COVID-19 on the basis of patients' acoustic signals. The main hypothesis of this proposal is that COVID-19 diagnosis and prognosis can be performed from the combination of information extracted from speech, breathing, and coughing sounds using a non-contact microphone.
Researcher: Dr. Yaron Orenstein, bioinformatics
Student: Mira Barshai
Detecting unique molecular structures in the Corona virus genome
The corona virus genome contains almost 30,000 nucleotides. This genome is not just a long string over A, C, G and U, but rather a structured molecule. The structure of the molecule, on top of its nucleotide content, can shed light on the function of different regions in the genome. Unfortunately, measuring the structure of the genome is a long, complex and time-consuming process. Instead, researchers often rely on measurements made on other genomes, train a model to predict them, and then apply them to novel genomes, such as the Corona virus genome.
In this project, we are looking for unique structures, called RNA G-quadruplexes, in the Corona virus genome. We plan to train a deep neural network to predict such structures based on a given RNA sequence. The network will be trained on available high-throughput data collected throughout the human transcriptome, providing millions of data points. Once we develop an accurate predictor, we will use it to predict RNA G-quadruplex structures in the Corona virus genome, and experimentally verify the top candidates. Verified structures may serve as potential therapeutic drug targets.